 Table of Contents
Table of Contents
 Previous Chapter
Previous Chapter
ACIS
The purpose of the Science Management Classes are to implement the top-level control of science data processing on the Back End Processor. This section discuses the overall control of science operations, and discuss the ScienceManager, ScienceMode, and ProcessMode classes.
Specific science modes (SmTimedExposure, SmContClocking), science data processing and the relevant classes are described in Section 37.0.
The following lists the uses of the Science Management classes. In practice, the first use listed covers each of the subsequent uses, which are individually listed to indicate each of the main steps needed to perform a science or bias run:
- Perform a bias or science run
- Initiate the setup of the hardware and software for a science or bias run
- Initiate the dump of parameters used by a science or bias run
- Initiate a bias computation for each of the selected CCDs
- Initiate image acquisition and processing for each of the selected CCDs
Figure 157 illustrates the overall relationships between the Science Manager and Science Mode classes.
FIGURE 157. Science Management Classes

ScienceManager - This class is a subclass of Executive::Task and is responsible for handling requests to start (startRun) and stop (stopRun) science and/or bias runs, and for directing a science mode through each stage of commanded run (goTaskEntry). This class has two types of functions, binding-functions and implementation functions. Binding functions are those functions which may be called from other tasks, to direct the science manager to take some sort of action (startRun, stopRun, notify, inhibit), or to query the state of the task (isIdle). Implementation functions are used internally by the science task, and are typically called by the task itself (requestEvent, waitForEvent, yield). The ScienceManager uses a passed ScienceMode to implement each stage of a bias or science run.
ScienceMode - This abstract class is responsible for providing top level functions for each stage of a science bias or data processing run. These functions include checking the integrity of a parameter block (checkBlock), setting up for the run (stageParameters, activateParameters, setup), dumping the parameters for the run (dumpParameters), determining if a bias computation is required by the mode's parameters (requiresBias), performing a bias computation (computeBias), loading bad pixel and column maps (loadBadMaps), processing science data (processData), stopping data processing (requestStop), and shutting down at the end of a run (terminate). It also provides a function used by the data processing classes, ProcessMode, to wait for telemetry packets (waitForPkt). Each instance of ScienceMode contains an array of pointers to ProcessMode instances, one for each Front End Processor in the instrument. Each referenced ProcessMode is used by the ScienceMode to handle science data records produced by a single Front End Processor. Not shown in the figure is the collection of utility member functions, used by the ScienceMode class, and its child classes, to implement the details of the run. The use of these functions is shown in Section 33.5, and are described in detail in Section 37.0.
SmTimedExposure - This is a subclass of ScienceMode and is responsible for configuring and performing a Timed Exposure bias or data processing run. This class is described in detail in Section 37.0. When configuring the ProcessMode pointers, this class uses subclasses of ProcessMode to handle different types of Timed Exposure data processing modes. For raw telemetry mode, it uses instances of the PmTeRaw class (not shown). For histogram mode, it uses PmHist instances (not shown). When processing events, SmTimedExposure uses instances of PmTeFaint3x3 for faint 3x3 event processing, PmTeFaintBias3x3 for faint-with-bias 3x3 event processing, and PmTeGraded for graded event telemetry production (not shown). A detailed description of the ProcessMode classes is provided in Section 37.0.
SmContClocking - This is a subclass of ScienceMode, and is responsible for configuring and performing a Continuous Clocking bias or data processing run. This class is described in detail in Section 37.0. When configuring the ProcessMode pointers, this class uses subclasses of ProcessMode to handle different types of Continuous Clocking data processing modes. For raw telemetry mode, it uses instances of the PmCcRaw class (not shown). When processing events, SmContClocking uses instances of PmCcFaint1x3 for faint 1x3 event processing, and PmCcGraded for graded event telemetry production (not shown). A detailed description of the ProcessMode classes is provided in Section 37.0.
ProcessMode - This abstract class is responsible for processing science data records, produced from a single Front End Processor. This class provides a top level function to process Front End Processor data records (processRecord). This class also provides a collection of member functions (not shown) used to configure the instance for a particular run. These include functions to indicate and retrieve which CCD is being processed (setCcdId, getCcdId), set and get the row and column scale factors and other image information (setGeometry, getGeometry, setSplitThreshold), set and get the ScienceMode from which to obtain telemetry packets (setMode, getMode), set and get the run's bias and data command and parameter block identifiers (setRunIdInfo, getRunIdInfo), and set and get the start times of the bias and data runs (setTimeBias, setTimeData, getTimeBias, getTimeData). This class also provides a set of common routines used by it and its subclasses. A detailed description of the ProcessMode classes is provided in Section 37.0.
Figure 158 illustrates the overall state behavior of the science manager class.
FIGURE 158. Science Manager state behavior
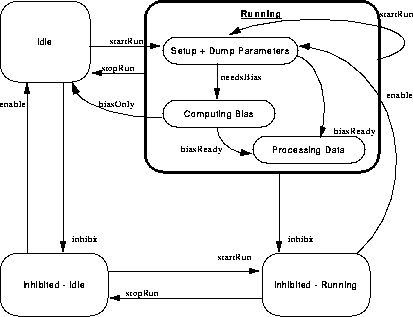
Initially, the science manager is idle. When it receives a startRun command, it initiates a new science run. If at any point in the process, the science manager receives a stopRun command, the run is terminated, and the manager goes idle. The startRun indicates whether or not this run should only compute bias, or should process data.
In general, startRun requests cause the science manager to start a science run, and stopRun requests cause the science manager to terminate the current run. A run may also be terminated once a "compute bias" completes, and no data processing was requested.
Whenever the science manager is inhibited, it maintains the desired running state while it remains inhibited, and once enabled, it either goes idle, or starts a run, depending on the last received startRun or stopRun command.
Figure 159 illustrates the overall steps used to conduct a bias and science run.
FIGURE 159. Bias and Science Run
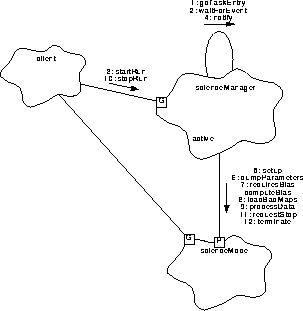
- Soon after startup, the executive invokes the main function of the Science Manager task, scienceManager.goTaskEntry(), which enters an infinite processing loop.
- goTaskEntry() waits for notification of a command by calling waitForEvent(). Whenever the taskMonitor queries the scienceManager, scienceManager wakes up from waitForEvent() and responds to the query using taskMonitor.respond() (not shown). This is continued until a command to perform a run is received.
- A client decides to start a science run. It calls scienceManager.startRun(), passing the command identifier initiating the run, the parameter block's slot identifier to use, the mode to run, scienceMode, and whether or not the run should only compute bias. For the purposes of this scenario, assume that both bias and data processing are to be performed.
- scienceManager.startRun() saves the passed information (such as the parameter block slot id, and whether or not this is a bias-only run), overwriting any pending requests (this occurrence is reported to software housekeeping, but is not shown nor described in this scenario), instructs the mode to copy the requested parameter blocks into its staging area using scienceMode.stageParameters() (not shown), and wakes up its task, using notify(). startRun then returns to its caller. Later, waitForEvent() returns, indicating that a request is present.
- goTaskEntry() copies the request information, tells the mode to activate the staged parameter blocks, using scienceMode.activateParameters() (not shown) and calls scienceMode.setup() to setup the hardware and software for the run.
- goTaskEntry() dumps the parameter blocks to telemetry, using scienceMode.dumpParameters() (see Section 33.5.3).
- goTaskEntry() tests if the parameters require a bias computation, using scienceMode.requiresBias(). If a bias is required by the parameter block, or if the startRun requested a bias-only run, goTaskEntry() calls scienceMode.computeBias() to compute the bias maps on all of the selected Front End Processors (see Section 33.5.4).
- goTaskEntry() loads the bad pixel or column maps into the Front End Processor's bias maps, using scienceMode.loadBadMaps(). NOTE: After adding pixels or columns from the bad pixel or column maps, or deleting the contents of a map, the ground should command a new bias calibration to be performed prior to processing any new science data.
- If startRun() did not request a bias-only computation, goTaskEntry() calls scienceMode.processData() to start data acquisition and processing (see Section 33.5.5). processData() does not return to goTaskEntry() until it is stopped by a call to scienceMode.requestStop().
- Later, while data acquisition and processing are being performed, the client tells the scienceManager to stop the run, using scienceManager.stopRun().
- stopRun() records the request. If a mode is active, stopRun() calls scienceMode.requestStop() to cause the mode to finish up its current data set and stop data processing.
- Once the data processing stops, scienceMode.processData() returns to goTaskEntry(). goTaskEntry() calls scienceMode.terminate() to cleanup from the processing and send a final run report to telemetry. goTaskEntry() then repeats its infinite loop, starting from step 2.
Figure 160 illustrates the high-level steps used to setup for a bias or science run.
FIGURE 160. Science/Bias Run Setup
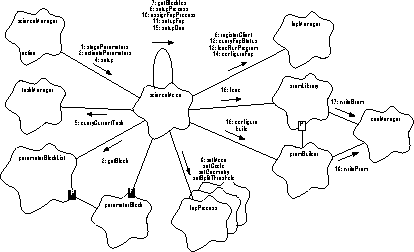
- When the scienceManager's binding function is instructed to start a new run, it tells the requested mode to stage its parameter blocks, using scienceMode.stageParameters().
- stageParameters() retrieves the requested parameter blocks from the parameter block list and saves them for use later, once the requested mode is ready to run, using parameterBlockList.getBlock().
- Later, once the manager's task is ready, it copies the request into its current state variables, and tells the mode to activate its staged parameter blocks, using scienceMode.activateParameters().
- The scienceManager then tells the current mode to prepare for processing, using scienceMode.setup().
- scienceMode.setup() asks the taskManager which task it is running under, using taskManager.queryCurrentTask().
- setup() tells the fepManager which task to notify, and with which events, using fepManager.registerClient().
- setup() then retrieves and stores the identifiers contained within the parameter blocks using getBlockIds().
- setup() calls setupProcess() to setup the data processing instances for the mode.
- setupProcess() configures each ProcessMode instance. It establishes itself as the source of telemetry packets, using fepProcess.setMode(). It associates the process with a CCD using fepProcess.setCcdId(), informs the process of the row and column scale factors, the row offset, and the output node configuration, using fepProcess.setGeometry(), and sets the split threshold levels, using fepProcess.setSplitThreshold().
- setupProcess() uses assignFepProcess() to install the configured ProcessMode's into the process pointer array.
- setup() calls setupFep() to load, run and configure each of the Front End Processors.
- setupFep() calls fepManager.queryFepStatus() to determine if the FEP is up and running, and to determine if a new bias computation is needed.
- If the FEP has power, but is not running, or if a customized Front End Processor code load is required, setupFep() calls fepManager.loadRunProgram() to install and run the default or custom FEP program.
- setupFep() uses fepManager.configureFep() to load the mode's FEP parameters into the running Front End Processors.
- setup() uses setupDea() to generate and load the Detector Electronics Assembly CCD Controller's sequencer images.
- setupDea() uses sramLibrary.load() to load the standard Sequencer RAM (SRAM) into one or more of the CCD controller sequencer memories.
- sramLibrary.load() uses deaManager.writeSram() to write to this RAM.
- setupDea() then uses the PRAM builder to construct and load the Program RAM into one or more of the CCD Controller's sequencer memories. The builder is configured using pramBuilder.configure(), and the PRAM program is built and loaded using pramBuilder.build().
- As the PRAM builder constructs the sequencer's program, it loads the program into sequencer memory using deaManager.writePram().
Figure 161 illustrates the steps involved in sending the parameter blocks to telemetry.
FIGURE 161. Parameter Dumps
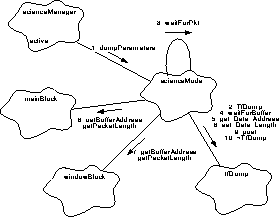
- The scienceManager instructs the setup mode to dump its parameter blocks, using scienceMode.dumpParameters().
- dumpParameters() declares a telemetry form instance, tfDump (TfDump()).
- dumpParameters() passes tfDump to waitForPkt() to wait for and associate a telemetry packet with the form.
- waitForPkt() uses tfDump.waitForBuffer() to wait for and allocate a telemetry packet buffer.
- dumpParameters() uses tfDump.get_Data_Address() to obtain the destination pointer for the parameter block within the telemetry packet buffer.
- It then uses mainBlock.getBufferAddress() to get the start of the parameter block buffer, and mainBlock.getPacketLength() to get the length of the block. It then copies the parameter block contents directly into the telemetry packet buffer.
- If a window list is used for the run, dumpParameters() gets the address and length of the list buffer using windowBlock.getBufferAddress() and windowBlock.getPacketLength(), and appends the window list to the main parameter block within the telemetry packet buffer.
- dumpParameters() then sets the data length in the telemetry packet using tfDump.set_Data_Length().
- dumpParameters() posts the packet buffer to telemetry, using tfDump.post().
- Once the packet buffer has been posed, dumpParameters() returns, causing the destructor for the form to be called (~TfDump).
Figure 162 illustrates how a bias computation is initiated.
FIGURE 162. Bias Computations
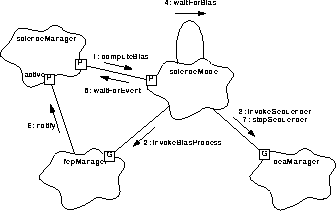
- scienceManager instructs the prepared mode (i.e. scienceMode.setup() must have already been called) to compute the CCD pixel bias values, using scienceMode.computeBias().
- computeBias() starts the bias computation process on each of the configured Front End Processors using fepManager.invokeBiasProcess().
- computeBias() starts the Detector Electronics Assembly CCD Controller Sequencers using deaManager.invokeSequencer(), and saves the microsecond science time-stamp, latched by the hardware when the command to start the sequencer was issued.
- computeBias() then waits for the bias computation to complete, using waitForBias().
- waitForBias() uses the current task to wait to be notified of the completion of the bias computation, using scienceManager.waitForEvent().
- Later, once all of the running Front End Processors have completed their respective bias computations, the fepManager notifies the installed client, using scienceManager.notify(), causing waitForEvent() to return.
- computeBias() stops the DEA's sequencers, using deaManager.stopSequencer(). It then calls loadBadMaps() (not shown) to load the bad pixel and columns into the bias map memory. computeBias() then calls useBiasThief() (not shown) to determine if the bias maps are to be telemetered. If so, computeBias() calls biasThief.biasReady() (not shown), causing the biasThief to send the maps to telemetry (see Section 33.5.1 for a description of the loading of the bad pixel and column maps).
Figure 163 illustrates the overall steps used to acquire and process CCD data
FIGURE 163. Data Acquisition and Processing
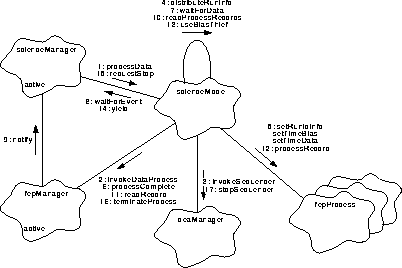
- scienceManager instructs the scienceMode to acquire and process data, using scienceMode.processData().
- processData() starts the Front End Processor's data processing routines, using fepManager.invokeDataProcess().
- processData() starts clocking all of the selected CCDs using deaManager.invokeSequencer(), which returns with the start time of the run. NOTE: All of the CCD sequencers start at the same time.
- processData() distributes the run and start time information to each ProcessMode using distributeRunInfo().
- distributeRunInfo() informs each ProcessMode of the command and parameter block identifiers for the most recent bias computation and for the current run using fepProcess.setRunIdInfo(). It informs each mode of the most recent bias computation start time using fepProcess.setTimeBias(), and of the start time of the current run using fepProcess.setTimeData().
- processData() enters its main processing loop, which terminates on an abort request, error, or when the Front End Processor indicates that processing is complete, via fepManager.processComplete().
- processData() waits for data to arrive from one or more of the Front End Processors, using waitForData().
- waitForData() uses the current task to wait for the event, scienceManager.waitForEvent().
- Later, one or more of the Front Ends reports that data is available. fepManager informs the installed client using scienceManager.notify().
- Once notified, waitForEvent() returns, and processData() calls readProcessRecords() to read and process one or more data records from one of the Front End Processors.
- readProcessRecords() gets one record from one of the Front Ends using fepManager.readRecord().
- If a record is returned by readRecord(), readProcessRecords() indexes the appropriate data processor, using the Front End identifier, and tells it to process the records, via fepProcess[fepId].processRecord(). In order to allow the biasThief task to run, readProcessRecords() repeats this for no more than 100 records, or until no more records are available (NOTE: The 100 figure can easily be patched).
- Once readProcessRecords() returns, processData() determines if the bias thief task is being used to trickle bias information to telemetry by calling useBiasThief().
- If the biases are being sent, processData() tells the current task to yield control to the bias processing task, using scienceManager.yield(). This happens for each set of FEP records processed by readProcessRecords(). After yielding, processData() then iterates the data processing loop (see Step 6).
- At some later time, the scienceManager can be commanded to stop the current run. It calls scienceMode.requestStop() to attempt to terminate the run. requestStop() then sets an internal flag indicating that a stop has been requested, and notifies the task that a stop has been requested, via scienceManager.notify() (not shown).
- Upon each iteration (Step 6 through Step 15) of the processing loop (starting with Step 6), processData() tests the state of the stop flag. If it is asserted, it tells the Front Ends to finish up their processing, using fepManager.terminateProcess(). processData() then continues its processing loop until fepManager.processComplete() indicates that all of the Front End's have finished.
- processData() then stops the CCD clocking, using deaManager.stopSequencer(), and returns.
Documentation:
- This class is responsible for managing science operations within the instrument, including storing parameter blocks, initiating science runs, and terminating science runs.
Export Control:
- Public
Cardinality:
- 1
Hierarchy:
Superclasses:
Task
Public Uses:
ScienceMode
Public Interface:
Operations:
ScienceManager()
goTaskEntry()
inhibit()
isIdle()
startRun()
stopRun()
Private Interface:
Has-A Relationships:
- ScienceMode* currentMode: This references the current science mode being run.
- Boolean currentBiasOnly: This variable indicates whether or not the current mode should only compute bias. If BoolTrue, only a bias computation is being performed. If BoolFalse, a science data run (with or without bias) is being run.
- unsigned currentCmdId: This is the command id which initiated the current run.
- ScienceMode* requestMode: This references the mode to be run.
- Boolean requestBiasOnly: This variable indicates whether the requested run should only compute bias.
- unsigned requestCmdId: This is the command id which is requesting the run.
- unsigned requestBlockId: This is the slot id of the parameter block to use for the requested run.
- DesiredState desiredState: This enumeration indicates what state the science manager should be in. It can have the following values:
SCIENCE_IDLE
SCIENCE_RUNNING
SCIENCE_INHIBITED_IDLE
SCIENCE_INHIBITED_RUN
Concurrency:
- Active
Persistence:
- Persistent
Public member of:
- ScienceManager
Arguments:
unsigned taskid
Documentation:
This is the constructor for the ScienceManager. taskid is the Nucleus RTX Task identifier for the science manager task. This function's initialization list first calls its parent's constructor, Task(), passing it taskid. It then initializes each of its instance variables.
Concurrency:
- Sequential
Public member of:
- ScienceManager
Return Class:
- void
Documentation:
- This is the main loop of the Science Manager Task. This function is responsible for waiting for state change requests, and responding to these requests.
Semantics:
- At the top of the FOREVER loop, if the desiredState is SCIENCE_RUNNING, use requestEvent() to acquire any pending state change or task query events, otherwise, use waitForEvent() to block until a such an event occurs. If a task query event is caught, respond using taskMonitor.respond(). If the desiredState is SCIENCE_RUNNING, copy the requestMode, requestCmdId and requestBiasOnly into the current variables, and then tell the desired mode to activate its staged parameters, using mode>activateParameters(). Then use mode->setup() to setup for a run, and mode>dumpParameters() to dump the parameter blocks for the run. If a bias is needed, use mode>computeBias() to compute the bias map. If data processing is needed, use mode>processData() to perform the data processing phase of the run. Once complete, use mode>terminate() to end the run and zero the current mode.
Concurrency:
- Synchronous
Public member of:
- ScienceManager
Return Class:
- void
Arguments:
Boolean on
Documentation:
This function controls whether or not science runs are inhibited (see Section 33.4). If on is BoolTrue, science runs are inhibited. If a run is in progress when the function is invoked, the current science run is terminated. New run requests are deferred until runs are enabled. If on is BoolFalse, then science runs are enabled. If a science run was terminated by an earlier call to inhibit(), or was deferred, the run is started.
Semantics:
- If on is BoolTrue: If desiredState is IDLE, set state to INHIBITED_IDLE. If desiredState is RUNNING, set state to INHIBITED_RUN. If a run is in progress, call mode->requestStop() to abort the run.
- If on is BoolFalse: If desiredState is INHIBITED_IDLE, set state to IDLE. If desiredState is INHIBITED_RUN, set state to RUNNING and notify the task that a run request is pending using notify().
Concurrency:
- Synchronous
Public member of:
- ScienceManager
Return Class:
- Boolean
Documentation:
- This function determines whether or not science processing is being performed. If there is no science processing in progress (currentMode is NULL, and desiredState is not RUNNING), it returns BoolTrue. If a science or bias run is in progress, it returns BoolFalse.
Concurrency:
- Synchronous
Public member of:
- ScienceManager
Return Class:
- Boolean
Arguments:
ScienceMode& mode
unsigned blockid
unsigned cmdid
Boolean biasOnly
Documentation:
This function starts a science run using the passed mode to process the setup, bias, run and shutdown states of the run. If biasOnly is BoolTrue, the manager will setup and compute the bias for the mode and stop. If biasOnly is BoolFalse, the manager will setup, compute the bias (if mandated by the parameter block), and proceed onto event processing until commanded to stop. blockid identifies which parameter slot to use for the run, and cmdid is the id of the command packet which is initiating the run.
Semantics:
- Copy mode, blockid, cmdid and biasOnly into the request parameters. Use mode.stageParameters() to load the parameter blocks into the mode's staging area. If the desiredState is inhibited, set it to INHIBITED_RUN to indicate that a run is being deferred, otherwise, set it to RUNNING. If a mode is already in progress, use currentMode>requestStop() to abort the active run. Use notify() to cause the task to check for the new run request.
Concurrency:
- Synchronous
Public member of:
- ScienceManager
Return Class:
- Boolean
Documentation:
- This function causes the Science Manager to stop the current science run. If no run is in progress, this function reports it to software housekeeping and returns BoolFalse. No further action is taken. If a run is underway, this function initiates termination of the run, and returns BoolTrue.
Semantics:
- If the desiredState is inhibited, set it to INHIBITED_IDLE, otherwise, set it to IDLE. If a mode is already in progress, use currentMode>requestStop() to end the active run.
Concurrency:
- Synchronous
Documentation:
- This abstract class represents a science processing mode.
Export Control:
- Public
Cardinality:
- n
Hierarchy:
Superclasses:
none
Implementation Uses:
FepManager
BiasThief
DeaManager
TlmForm
Public Interface:
Operations:
ScienceMode()
activateParameters()
checkBlock()
computeBias()
dumpParameters()
loadBadMaps()
processData()
requestStop()
requiresBias()
setup()
stageParameters()
terminate()
waitForPkt()
Protected Interface:
Has-A Relationships:
- Task* curTask: This variable points to the task under which the science mode is running. This variable is assigned during the call to setup().
- FilterGrade filterGrade[6]: This is a set of filters which select events based on their respective grade codes. See Section 37.15 for a description of this class.
- FilterPh filterPh[6]: This is a set of filters which select events based on their corrected amplitude. See Section 37.16 for a description of this class.
- FilterWindow filterWindow[6]: This is a set of filters which select events or raw pixels based on their position within the CCD image. See Section 37.14 for a description of this class.
Operations:
assignFepProcess()
distributeRunInfo()
getBlockIds()
getFepRequestType()
readProcessRecords()
setupDea()
setupFep()
setupProcess()
useBiasThief()
waitForBias()
waitForData()
waitForEvent()
Private Interface:
Has-A Relationships:
- unsigned startTimeBias: This variable contains a copy of the science microsecond timestamp at the time the DEA sequencer was started for a bias computation.
- unsigned startTimeData: This variable contains a copy of the science microsecond time stamp latched when the DEA sequencers were started for science data processing.
- ProcessMode* fepProcess[6]: This array of ProcessMode pointers is indexed by Front End Processor id. Each pointer points to the ProcessMode responsible for the corresponding FEP. If 0, then the corresponding FEP is not in-use. This array is set during the call to setup().
- Boolean stopRequest: This variable indicates whether a stop run has been requested. If BoolTrue, then the mode will complete the current exposure set and end data processing.
- unsigned cmdIdBias: This is the id of the command which caused the most recent bias to be computed. NOTE: When executing a bias calibration, this value is identical to cmdIdCurrent.
- unsigned blockIdBias: This is the id of the parameter block used to perform the most recent bias computation. NOTE: When executing a bias calibration, this value is identical to blockIdCurrent.
- unsigned cmdIdCurrent: This is the id of the command which invoked the current run.
- unsigned blockIdCurrent: This is the id of the parameter block used to configure the current run.
- unsigned winIdCurrent: This is the id of the window parameter block used for the current run.
- RINGREC record: This buffers one FEP to BEP data record.
- unsigned pktPollTimeout: This is the number of ticks to wait for a telemetry packet buffer between checks for abort commands and responses to the task monitor.
Concurrency:
- Guarded
Persistence:
- Persistent
Protected member of:
- ScienceMode
Documentation:
- This is the constructor for the ScienceMode. The initialization statements first set each of the mode's instance variables, setting startTimeData to 0xffffffff. The body of the constructor then writes 0 into each pointer in the fepProcess array.
Concurrency:
- Guarded
Public member of:
- ScienceMode
Return Class:
- void
Documentation:
- This function copies the staged parameter block and window list parameter block into the mode's active copies. This function must be implemented by all subclasses of this class.
Concurrency:
- Guarded
Protected member of:
- ScienceMode
Return Class:
- void
Arguments:
FepId fepId
ProcessMode* process
Documentation:
This function installs a FEP process mode, process, into the processor slot associated with the FEP, fepId.
Concurrency:
- Guarded
Public member of:
- ScienceMode
Return Class:
- Boolean
Arguments:
unsigned blockid
Documentation:
This function acquires and verifies the parameter block identified by blockid. If the parameter block id is valid, and the indexed block is intact, this function returns BoolTrue. If the parameter block id is invalid, or if the parameter block has been corrupted, this function returns BoolFalse. This function must be implemented by each subclass of this class.
Concurrency:
- Synchronous
Public member of:
- ScienceMode
Return Class:
- Boolean
Documentation:
- This function causes the science mode to compute the CCD bias values for all configured CCDs, and load the bad pixel and column maps into the FEP's memory. Calling this function will start clocking the DEA sequencers, and will cause the FEPs to modify their bias maps. If configured to do so, this function will also cause the BiasThief task to start sending bias information as soon as it becomes available.
Preconditions:
- setup() must be called prior to calling this function.
Postconditions:
- Upon successful initiation of a bias computation, cmdIdBias, blockIdBias, and startTimeBias will contain, respectively, the id of the current command, of the current parameter block, and the microsecond timestamp sampled when the DEA was commanded to start clocking the CCDs for the bias computation.
Concurrency:
- Guarded
Protected member of:
- ScienceMode
Return Class:
- void
Documentation:
- This function distributes the command and parameter block identifiers, and data processing and bias computation start times to each active fepProcess.
Preconditions:
- setup() must have been called prior to calling this function
Concurrency:
- Guarded
Public member of:
- ScienceMode
Return Class:
- void
Documentation:
- This function dumps the mode's parameter blocks to telemetry. Each subclass must implement this function.
Preconditions:
- setup() must be called prior to calling this function.
Concurrency:
- Guarded
Protected member of:
- ScienceMode
Return Class:
- void
Arguments:
unsigned& blockId
unsigned& winId
Documentation:
This function returns the identifiers contained within the active parameter block and window list parameter block. On return, blockId contains the parameter block identifier contained within the main parameter block. If a window list was specified, winId, this function stores the identifier contained within the window list block. If no window list is active, the function stores 0xffffffff in the winId. Each subclass of this class must implement this function.
Preconditions:
- setup() must have been called prior to calling this function
Concurrency:
- Guarded
Protected member of:
- ScienceMode
Return Class:
- int
Documentation:
- This function returns the request type to use when commanding the FEPs to start a run. Each subclass of this class must supply this function.
Concurrency:
- Synchronous
Public member of:
- ScienceMode
Return Class:
- void
Documentation:
- This function loads the bad pixel and column maps into the configured Front End Processor's pixel bias memory and adjusts the bias map parity bits associated with each affected memory location. Each subclass is required to implement this function.
- NOTE: The address of the bias memory is fixed, however, the address of the pixel parity plane must be obtained from the code running on the FEP via a query to the FEP during the call to setup().
Preconditions:
- setup() must be called prior to calling this function. If computeBias() is to be called for this run, it must be called prior to calling this function.
Postconditions:
- The FEP's pixel bias map values corresponding to bad pixels or columns will be overwritten with codes flagging them as bad. To remove these flags, computeBias() must be re-invoked.
Concurrency:
- Guarded
Public member of:
- ScienceMode
Return Class:
- Boolean
Documentation:
- This function causes the science mode to acquire and process event or pixel data from the FEPs. This function returns only if the run is aborted, an error occurs, or requestStop() is called. This function returns BoolTrue if data acquisition and processing was started, and BoolFalse if an error prevented data processing.
Preconditions:
- setup() must be called prior to calling this function.
Semantics:
- Start data processing on the configured FEPs using fepManager.invokeDataProcess(). Then start the CCD sequencers using deaManager.invokeSequencer(), obtaining the science microsecond time-stamp for the run. Then distribute the start-times to each of the data processing modes using distributeRunInfo().
- Enter the processing loop. The loop terminates on error, or when fepManager.processComplete() indicates that the FEPs are done producing data. In the loop body, check stopRequest. If it is set, call fepManager.terminateProcess() (once the FEPs are done, fepManger.processComplete() will return BoolTrue). Call waitForData() to let other tasks run while waiting for one or more of the FEPs to produce data. Read and process records from one or more of the FEPs using readProcessRecords(). Finally, check if the bias thief is in use, and yield if so.
- Once the loop terminates, call deaManager.stopSequencer() to stop clocking images.
Concurrency:
- Guarded
Protected member of:
- ScienceMode
Return Class:
- Boolean
Documentation:
- This function reads and processes a series of records from one or more of the active FEPs. The function returns BoolTrue is successful, and BoolFalse on abort request or fatal error.
Preconditions:
- This function is intended to be called only during data processing.
Concurrency:
- Guarded
Public member of:
- ScienceMode
Return Class:
- void
Documentation:
- This function signals the mode to stop data processing. It sets stopRequest to BoolTrue. If curTask is not 0, it notifies it that a stop request has been issued.
Concurrency:
- Synchronous
Public member of:
- ScienceMode
Return Class:
- Boolean
Documentation:
- This function determines whether or not the configured mode requires a bias computation prior to data processing using the mode's parameter block. If a fresh bias computation is required, it returns BoolTrue. If a fresh bias computation is not indicated by the block, it returns BoolFalse. Each subclass must implement this function.
Concurrency:
- Guarded
Public member of:
- ScienceMode
Return Class:
- Boolean
Arguments:
unsigned blockid
unsigned cmdid
Documentation:
This function sets up all hardware and software needed to execute a science run or bias computation. blockid is the parameter block slot identifier to use for the run. cmdid is the id of the command causing the mode to be run.
Concurrency:
- Guarded
Protected member of:
- ScienceMode
Return Class:
- void
Documentation:
- This function sets up the Detector Electronics Assembly for the current mode. All child classes must implement this function.
Concurrency:
- Guarded
Protected member of:
- ScienceMode
Return Class:
- void
Documentation:
- This function loads and configures the Front End Processors for the mode. All subclasses must implement this function.
Concurrency:
- Guarded
Protected member of:
- ScienceMode
Return Class:
- void
Documentation:
- This function sets up the ProcessMode instances, for the current run. Each subclass must implement this function.
Concurrency:
- Guarded
Public member of:
- ScienceMode
Return Class:
- void
Arguments:
unsigned blockid
Documentation:
This function makes a local copy of the parameter block identified by blockid, and any referenced parameter blocks, in preparation for being commanded to use the blocks later. Each subclass is required to implement this function.
Concurrency:
- Guarded
Public member of:
- ScienceMode
Return Class:
- void
Documentation:
- This function causes the science mode to complete its bias/processing operations, perform a post-run integrity check, and telemeter a summary of the run. Each subclass is required to implement this function.
Concurrency:
- Guarded
Protected member of:
- ScienceMode
Return Class:
- Boolean
Documentation:
- This function determines whether or not to use the Bias Thief task to telemeter the contents of the FEP's Bias Map memory. It returns BoolTrue if the task should be configured and notified when the maps are ready to be sent, and BoolFalse if the bias thief is not used for the current mode. Each subclass is required to implement this function.
Preconditions:
- setup() must be called prior to calling this function.
Concurrency:
- Guarded
Protected member of:
- ScienceMode
Return Class:
- Boolean
Documentation:
- This function suspends the current task until all of the biases have been computed on all of the Front End Processors, or until a command is issued to the mode. This function also periodically wakes up and responds to queries from the taskMonitor. It returns BoolTrue if the bias computation completed, and BoolFalse on an abort or error.
Preconditions:
- A bias computation must have been started on the FEPs prior to calling this function.
Concurrency:
- Guarded
Protected member of:
- ScienceMode
Return Class:
- Boolean
Documentation:
- This function waits for notification that data is available from at least one of the enabled FEPs. This function also responds to queries from the taskMonitor. The function returns BoolTrue if data is available, and BoolFalse on error, or if the run has been aborted.
Preconditions:
- Data processing must be active on the FEPs.
Concurrency:
- Guarded
Protected member of:
- ScienceMode
Return Class:
- Boolean
Arguments:
unsigned event
Documentation:
This function waits for the event (s) indicated by event, while also responding to taskMonitor queries, and checking for errors and abort requests. If event is received with no errors, the function returns BoolTrue. If the run was aborted, or an error occurs, the function returns BoolFalse.
Concurrency:
- Guarded
Public member of:
- ScienceMode
Return Class:
- Boolean
Arguments:
TlmForm& form
Documentation:
This function instructs the telemetry form to obtain a telemetry packet buffer for the form. This function periodically checks for task monitor queries and commanded state changes. If a packet is obtained, the function returns BoolTrue. If a command or error has aborted the run, the function returns BoolFalse.
Concurrency:
- Guarded
Documentation:
- This class represents a data processing mode for a single FEP.
Export Control:
- Public
Cardinality:
- n
Hierarchy:
Superclasses:
none
Public Uses:
TlmForm
ScienceMode
Public Interface:
Operations:
ProcessMode()
getCcdId()
getFepId()
getGeometry()
getMode()
getRunIdInfo()
getTimeBias()
getTimeData()
processRecord()
setCcdId()
setFepId()
setGeometry()
setMode()
setRunIdInfo()
setSplitThreshold()
setTimeBias()
setTimeData()
Protected Interface:
Has-A Relationships:
- EventExposure expInfo: This contains information about the current exposure and run. Its run information, such as the configured split thresholds and image scale and offset factors, is loaded by the call to setRunInfo. The function processRecord loads exposure-varying information, such as the overclock delta values, by passing FEP produced exposure start and end records to this object's member functions.
Operations:
getExposureInfo()
waitForPkt()
Private Interface:
Has-A Relationships:
- CcdId ccdid: This is the CCD being processed by this instance.
- FepId fepid: This is the FEP being processed by this instance.
- ScienceMode* modePtr: This variable points to the science mode using the event processor.
- unsigned runCmdIdData: This variable contains the command id used to start the science run.
- unsigned runCmdIdBias: This variable contains the command id which initiated the most recent bias computation.
- unsigned runPblockData: This is the parameter block identifier used to configure the current run.
- unsigned runPblockWin: This is the window parameter block identifier being used for the current run.
- unsigned runPblockBias: This is the parameter block identifier used to configure the most recent bias computation.
- unsigned runTimeBias: This is the start time (latched BEP microsecond science timestamp) of the most recent bias computation.
- unsigned runTimeData: This is the start time (latched BEP microsecond science timestamp) of the current run.
Concurrency:
- Guarded
Public member of:
- ProcessMode
Documentation:
- This is the constructor for the ProcessMode class. Its initialization statements zero all of its instance variables.
Concurrency:
- Sequential
Public member of:
- ProcessMode
Return Class:
- CcdId
Documentation:
- This function returns the CCD identifier being handled by this instance.
Preconditions:
- setCcdId() must be called prior to using this function.
Concurrency:
- Synchronous
Public member of:
- ProcessMode
Return Class:
- FepId
Documentation:
- This function returns the FEP identifier being handled by this instance.
Preconditions:
- setFepId() must be called prior to using this function.
Concurrency:
- Synchronous
Public member of:
- ProcessMode
Return Class:
- void
Arguments:
unsigned& rowscale
unsigned& colscale
unsigned& rowoffset
QuadMode& nodeSelect
Documentation:
This function supplies the row and column scale factors, and row offset factor reported via setGeometry. rowscale is the number of summed rows per pixel row, colscale is the number of summed columns per pixel, and rowoffset is the number of skipped CCD rows. nodeSelect indicates which output node configuration is in use (Full, Diagnostic, AC, or BD).
Preconditions:
- setGeometry() must be called prior to using this function.
Concurrency:
- Synchronous
Public member of:
- ProcessMode
Return Class:
- ScienceMode*
Documentation:
- This function returns a pointer to the science mode invoking this data process.
Preconditions:
- setMode() must be called prior to using this function.
Concurrency:
- Synchronous
Public member of:
- ProcessMode
Return Class:
- void
Arguments:
unsigned& dataCmdId
unsigned& dataBlockId
unsigned& dataWinId
unsigned& biasCmdId
unsigned& biasBlockId
Documentation:
This function returns the identification information used for the current data process. Upon returning, dataCmdId will be set to the id of the command which started the run, dataBlockId and dataWinId will be set to the parameter block id and window parameter block id used to configure the run. biasCmdId will be set to the id of the command which initiated the most recent bias computation, and biasBlockId is the parameter block id used to configure that computation.
Preconditions:
- setRunIdInfo() must be called prior to calling this function.
Concurrency:
- Synchronous
Public member of:
- ProcessMode
Return Class:
- unsigned
Documentation:
- This function returns the start time (the latched BEP microsecond science timestamp) of the most recent bias computation.
Preconditions:
- setTimeBias() must be called prior to using this function.
Concurrency:
- Synchronous
Public member of:
- ProcessMode
Return Class:
- unsigned
Documentation:
- This function returns the start time (the latched BEP microsecond science timestamp) of the current science data run.
Semantics:
- setTimeData() must be called prior to using this function.
Concurrency:
- Synchronous
Public member of:
- ProcessMode
Return Class:
- Boolean
Arguments:
const RINGREC& record
Documentation:
This function processes one FEP block buffer filled with a science record produced by a Front End Processor. record contains a copy of the FEP record block. If a run-terminating error occurs, or the run is aborted, this function returns BoolFalse. If the records were successfully parsed, or ignored, it returns BoolTrue. Each subclass replaces this member function with their own mode-specific code, and will explicitly call this function for records which they don't handle.
Concurrency:
- Guarded
Public member of:
- ProcessMode
Return Class:
- void
Arguments:
CcdId ccdId
Documentation:
This function associates this instance with a particular CCD, ccdId
Concurrency:
Guarded
Public member of:
- ProcessMode
Return Class:
- void
Arguments:
FepId fep
Documentation:
This function associates this instance with a particular FEP, fep.
Concurrency:
- Guarded
Public member of:
- ProcessMode
Return Class:
- void
Arguments:
unsigned rowscale
unsigned colscale
unsigned rowoffset
QuadMode nodeSelect
Documentation:
This function sets the scaling and offset factors for the image being clocked out of the CCD. rowscale is the number of summed CCD rows per produced pixel row. colscale is the number of CCD columns per produced pixel. rowoffset is CCD row index of the first processed CCD row. nodeSelect indicates which output node configuration is in use (Full, Diagnostic, AC, or BD).
Concurrency:
- Guarded
Public member of:
- ProcessMode
Return Class:
- void
Arguments:
ScienceMode* mode
Documentation:
This function associates the science mode instance, mode, with this processing instance.
Concurrency:
- Guarded
Public member of:
- ProcessMode
Return Class:
- void
Arguments:
unsigned dataCmdId
unsigned dataBlockId
unsigned dataWinId
unsigned biasCmdId
unsigned biasBlockId
Documentation:
This function sets the run identification information for the process. dataCmdId is the id of the command which initiated the data process, dataBlockId is the parameter block id used to configure the run, and dataWinId is the parameter block id of the window list used to filter the data. biasCmdId is the id of the command which initiated the most recent bias computation and biasBlockId is the id of the parameter block used to configure the most recent bias computation.
Concurrency:
- Guarded
Public member of:
- ProcessMode
Return Class:
- void
Arguments:
unsigned biasTime
Documentation:
This function stores the start time (the latched BEP microsecond science timestamp) of the most recent bias computation, biasTime.
Concurrency:
- Guarded
Public member of:
- ProcessMode
Return Class:
- void
Arguments:
unsigned dataTime
Documentation:
This function stores the start time (the latched BEP microsecond science timestamp) of the current science data run, dataTime.
Concurrency:
- Guarded
Protected member of:
- ProcessMode
Return Class:
- Boolean
Arguments:
TlmForm& form
Documentation:
This function waits tells the form to acquire a telemetry packet buffer, using the associated science mode, mode.
Concurrency:
- Guarded
 Table of Contents
Table of Contents
 Next Chapter
Next Chapter
 Table of Contents
Table of Contents
 Previous Chapter
Previous Chapter
 Table of Contents
Table of Contents
 Previous Chapter
Previous Chapter