
Next: 3.2 Commanding Up: 3.1.5 Bias Determination Previous: 3.1.5 Bias Determination
![]()
![]()
![]()
![]()

Next: 3.2 Commanding
Up: 3.1.5 Bias Determination
Previous: 3.1.5 Bias Determination
Ford and Somigliana at MIT have investigated a class of algorithms that possesses the considerable advantage that they use no additional FEP memory beyond the Image and Bias Map Buffers and a modest amount of Data Cache. At any moment during the course of the algorithm, each pixel is represented by only two quantities - the 12-bit value in the Image map which will be overwritten by each fresh exposure, and the 12-bit value in the Bias Map whose value must converge to the desired bias threshold level.
The distribution of values, pi, of a pixel observed N times (![]() ) may be approximated by a narrow Gaussian,
) may be approximated by a narrow Gaussian, ![]() , with the addition of a few outlying
values, especially those with
, with the addition of a few outlying
values, especially those with ![]() produced by X-rays or
ionizing particles. By definition, p0 is the modal value and
produced by X-rays or
ionizing particles. By definition, p0 is the modal value and
![]() is the width, typically a few data units. For new
CCDs, not yet subjected to radiation damage, both p0 and
is the width, typically a few data units. For new
CCDs, not yet subjected to radiation damage, both p0 and ![]() are nearly identical from pixel to pixel, except for a few pathological
`hot' or `flickering' pixels.
are nearly identical from pixel to pixel, except for a few pathological
`hot' or `flickering' pixels.
When a CCD is damaged by radiation, it is anticipated that p0 and
![]() will increase with time, and at rates that vary randomly from
pixel to pixel. Such behavior has been reported by Andy Rasmussen from
a study of the CCDs aboard the ASCA observatory. Algorithms that use
no additional storage are unable to estimate
will increase with time, and at rates that vary randomly from
pixel to pixel. Such behavior has been reported by Andy Rasmussen from
a study of the CCDs aboard the ASCA observatory. Algorithms that use
no additional storage are unable to estimate ![]() from pi, so
they concentrate on the modal value p0. Once this has been
computed for all pixels, its variance
from pi, so
they concentrate on the modal value p0. Once this has been
computed for all pixels, its variance ![]() serves as a good approximation to
serves as a good approximation to ![]() .
.
The algorithms start by examining a series of exposures to `condition' the pi values and derive bi, an estimate of p0. After the first exposure, the best estimate of b1 is clearly p1. After the i'th exposure, two possible algorithms yield improved values of bi, viz.
| |
(5) |
| |
(6) |
| |
(7) |
After N conditioning exposures, the Bias Map Buffer contains values
bN that are guaranteed to lie within a few ![]() 's of the modal
values p0. These can now be used as rejection thresholds to
identify X-ray and ionizing particle events in subsequent calibration
exposures, and thereby improve their own values in the process. This
is achieved by making a further set of M exposures, i.e. 0 < N < i <
(N+M). The new pixel values pi are subjected to the following
two-step algorithm:
's of the modal
values p0. These can now be used as rejection thresholds to
identify X-ray and ionizing particle events in subsequent calibration
exposures, and thereby improve their own values in the process. This
is achieved by making a further set of M exposures, i.e. 0 < N < i <
(N+M). The new pixel values pi are subjected to the following
two-step algorithm:
| (8) |
In the second step, the Image Map pixels are re-examined and those with values less than 4095 are used to refine the Bias Map values,
| bi = Cpi + (1 - C)bi-1 | (9) |
The principal advantage of this class of algorithms is that they are very simple to implement, requiring just a few lines of computer code. Preliminary tests show that the resulting bias maps compare favorably with those generated from `strip-by-strip' algorithms. However, they do have some weaknesses, as follows:
MEAN-I2L2
This algorithm calculates the mean and ![]() of N values of a pixel. It then rejects values that diverge from the
mean by more than
of N values of a pixel. It then rejects values that diverge from the
mean by more than ![]() , and then recalculates the mean and
, and then recalculates the mean and
![]() .
.
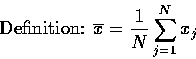 |
(10) |
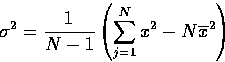 |
(11) |
| (12) |
The algorithm is expected to work best with Gaussian or symmetric heavy-tailed distributions in the presence of outlier points for which the assumption of normality does not hold. These outlier points are rejected and the remaining data are treated as Gaussian.
The calculation is accelerated by saving the intermediate values ![]() and
and ![]() . During the rejection phase, rejected
values are subtracted from
. During the rejection phase, rejected
values are subtracted from ![]() and
and ![]() and the
sample number is updated.
and the
sample number is updated.
MEDIAN
The algorithm calculates the median of N pixel
values. It sorts them in ascending order, and identifies the central
value, thereby automatically rejecting the outliers which will be
sorted to one end of the list or the other. Once the {xn} are
sorted, the formula for the median is:
![\begin{displaymath}
x_{med} = \left\{ \begin{array}
{ll}
x_{\frac{(N + 1)}{2}} ...
...\frac{N}{2} + 1 \right]}) &
\mbox{N even} \end{array} \right.\end{displaymath}](img95.gif) |
(13) |
| (14) |
A value of k = 3 can be considered to be a good approximation to reject severe outliers. The termination rule for the median is defined as follows:
MEDMEAN
This algorithm calculates the median value after excluding
some of the highest and lowest values, and excluding values
based on the variance about a trial mean. Specifically, it
takes the N values for a specific pixel pi, i=0,N-1,
where N is the number of conditioning exposures, and removes
the L largest values and the M smallest values.
Using the remaining N-L-M values, it computes the median
value, ![]() , and the variance,
, and the variance, ![]() , of
those pi less than
, of
those pi less than ![]() :
:
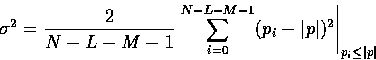 |
(15) |
The trial set of pi are compared to the trial ![]() ,and any values which do not fall within
,and any values which do not fall within ![]() are removed,
i.e., any pi which do not satisfy the condition:
are removed,
i.e., any pi which do not satisfy the condition:
| |
(16) |
Finally, the bias value is computed based on the remaining pi, by the normal equation for the mean:
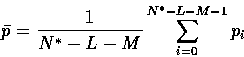 |
(17) |

John Nousek